[1] "patient_ID"
[2] "date_of_diagnosis"
[3] "method_of_detection"
[4] "date_of_birth"
[5] "age_at_diagnosis"
[6] "age_category"
[7] "gender"
[8] "height"
[9] "weight"
[10] "BMI"
[11] "BMI_category"
[12] "menopausal_status"
[13] "body_surface_area"
[14] "smoking"
[15] "alcohol_abuse"
[16] "hypertension"
[17] "hyperlipidemia"
[18] "diabetes"
[19] "comorbidities"
[20] "age_menarche"
[21] "oral_anticonceptive_use"
[22] "oral_anticonceptive_duration"
[23] "fertility_treatment"
[24] "age_first_pregnancy"
[25] "age_last_pregnancy"
[26] "pregnancy_A"
[27] "pregnancy_P"
[28] "pregnancy_G"
[29] "Age.FFTP"
[30] "Interval.1st.FTP"
[31] "breast_feeding"
[32] "breast_feeding_duration"
[33] "age_menopause"
[34] "hormone_replacement"
[35] "familial_history_breast_ovary"
[36] "familial_history_breast_ovary_line"
[37] "germline_mutation_testing_performed"
[38] "germline_mutation_testing_year_most_recent_test"
[39] "germline_mutation_testing_result"
[40] "visible_on_mammogram"
[41] "diameter_mammogram_at_diagnosis"
[42] "number_of_suspected_foci_mammogram"
[43] "breast_density_score_mammogram"
[44] "diameter_ultrasound_at_diagnosis"
[45] "Number_of_adenopathies_expected_on_ultrasound"
[46] "MRI_breast_performed"
[47] "diameter_MRI_at_diagnosis"
[48] "number_of_suspected_foci_MRI"
[49] "breast_density_score_MRI"
[50] "Number_of_adenopathies_expected_on_MRI_breast"
[51] "Number_of_adenopathies_expected_on_other_imaging"
[52] "primary_date_of_histological_diagnosis"
[53] "primary_laterality"
[54] "TNM_cT_at_diagnosis"
[55] "TNM_cN_at_diagnosis"
[56] "TNM_cM_at_diagnosis"
[57] "diameter_radiology_at_diagnosis"
[58] "tumor_grade_biopsy"
[59] "ER_Allred_biopsy"
[60] "ER_H_score_biopsy"
[61] "PR_Allred_biopsy"
[62] "PR_H_score_biopsy"
[63] "HER2_IHC_score_biopsy"
[64] "HER2_FISH_biopsy"
[65] "HER2_ratio_biopsy"
[66] "Ki67_biopsy"
[67] "ER_Allred_biopsy_2nd_lesion"
[68] "ER_H_score_biopsy_2nd_lesion"
[69] "PR_Allred_biopsy_2nd_lesion"
[70] "PR_H_score_biopsy_2nd_lesion"
[71] "HER2_IHC_score_biopsy_2nd_lesion"
[72] "HER2_FISH_biopsy_2nd_lesion"
[73] "HER2_ratio_biopsy_2nd_lesion"
[74] "Ki67_biopsy_2nd_lesion"
[75] "number_of_suspected_foci"
[76] "Number_of_adenopathies_expected_on_imaging"
[77] "neo_adjuvant_therapy"
[78] "TNM_ycT_after_neo_adjuvant_therapy"
[79] "TNM_ycN_after_neo_adjuvant_therapy"
[80] "neo_adjuvant_therapy_start_date"
[81] "neo_adjuvant_chemotherapy_scheme"
[82] "neo_adjuvant_chemotherapy_BSA_used"
[83] "neo_adjuvant_chemotherapy_BSA_capping"
[84] "neo_adjuvant_chemotherapy_completion"
[85] "neo_adjuvant_HER2_therapy_scheme"
[86] "neo_adjuvant_endocrinetherapy_scheme"
[87] "neo_adjuvant_endocrinetherapy_duration"
[88] "neo_adjuvant_other"
[89] "neo_adjuvant_therapy_end_date"
[90] "surgery_date"
[91] "surgery_type_breast"
[92] "surgery_type_axilla"
[93] "TNM_pT_resection_specimen"
[94] "TNM_pN_resection_specimen"
[95] "diameter_pathology_resection_specimen"
[96] "residual_tumorbed"
[97] "number_of_foci_resection_specimen"
[98] "multifocality"
[99] "tumor_grade_resection_specimen"
[100] "resection_margin_resection_specimen"
[101] "ER_Interpretation"
[102] "PR_Interpretation"
[103] "HER2_Interpretation"
[104] "Ki67_resection_specimen"
[105] "presence_DCIS_resection_specimen"
[106] "presence_LCIS_resection_specimen"
[107] "total_ALN_removed"
[108] "positive_ALN"
[109] "Micro_vs_macrometastases"
[110] "ALN_maxdiameter"
[111] "lobular_subtype"
[112] "ER_Allred_resection_specimen"
[113] "ER_H_score_resection_specimen"
[114] "PR_Allred_resection_specimen"
[115] "PR_H_score_resection_specimen"
[116] "HER2_IHC_score_resection_specimen"
[117] "HER2_FISH_resection_specimen"
[118] "HER2_ratio_resection_specimen"
[119] "ER_Allred_resection_specimen_2nd_lesion"
[120] "ER_H_score_resection_specimen_2nd_lesion"
[121] "PR_Allred_resection_specimen_2nd_lesion"
[122] "PR_H_score_resection_specimen_2nd_lesion"
[123] "HER2_IHC_score_resection_specimen_2nd_lesion"
[124] "HER2_FISH_resection_specimen_2nd_lesion"
[125] "HER2_ratio_resection_specimen_2nd_lesion"
[126] "Ki67_resection_specimen_2nd_lesion"
[127] "E.cadherin"
[128] "Antibody_E.cadherin"
[129] "B.catenin"
[130] "p120_catenin"
[131] "lymphatic_invasion_resection_specimen"
[132] "radiotherapy"
[133] "GEP_type"
[134] "GEP_outcome"
[135] "adjuvant_chemotherapy"
[136] "adjuvant_chemotherapy_scheme"
[137] "adjuvant_HER2"
[138] "adjuvant_HER2_scheme"
[139] "adjuvant_endocrinetherapy"
[140] "adjuvant_endocrinetherapy_scheme1"
[141] "adjuvant_endocrinetherapy_scheme1_duration"
[142] "adjuvant_endocrinetherapy_scheme2"
[143] "adjuvant_endocrinetherapy_scheme2_duration"
[144] "Total_duration_endocrine_treatment"
[145] "adjuvant_other"
[146] "meta_brain_nonleptomeningeal_first_metastases"
[147] "Meta_leptomeningeal_first_metastases"
[148] "meta_bones_first_metastases"
[149] "meta_skin_first_metastases"
[150] "meta_lungs_first_metastases"
[151] "meta_liver_first_metastases"
[152] "meta_abdomen_extrahepatic_first_metastases"
[153] "meta_reproductive_organs_first_metastases"
[154] "meta_lymph_nodes_first_metastases"
[155] "meta_other_first_metastases"
[156] "Systemic_treatment_firstline"
[157] "surgery_1st_line_metastatic"
[158] "radiotherapy_1st_line_metastatic"
[159] "chemotherapy_1st_line_metastatic"
[160] "HER2_1st_line_metastatic"
[161] "endocrinetherapy_1st_line_metastatic"
[162] "treatment_1st_line_other_metastatic"
[163] "treatment_reduction_1st_line_metastatic"
[164] "clinical_response_1st_line_metastatic"
[165] "first_progression_distant_disease_metastatic"
[166] "date_first_progression_metastatic"
[167] "meta_brain_nonleptomeningeal_atfirstprogression"
[168] "meta_leptomeningeal_atfirstprogression"
[169] "meta_bones_atfirstprogression"
[170] "meta_skin_atfirstprogression"
[171] "meta_lungs_atfirstprogression"
[172] "meta_liver_atfirstprogression"
[173] "meta_abdomen_extrahepatic_atfirstprogression"
[174] "meta_reproductive_organs_atfirstprogression"
[175] "meta_lymph_nodes_atfirstprogression"
[176] "meta_other_atfirstprogression"
[177] "systemic_treatment_secondline"
[178] "radiotherapy_2nd_line_metastatic"
[179] "chemotherapy_2nd_line_metastatic"
[180] "HER2_2nd_line_metastatic"
[181] "endocrinetherapy_2nd_line_metastatic"
[182] "treatment_2nd_line_other_metastatic"
[183] "treatment_reduction_2nd_line_metastatic"
[184] "clinical_response_2nd_line_metastatic"
[185] "second_progression_distant_disease_metastatic"
[186] "date_second_progression_metastatic"
[187] "number_of_lines_metastatic"
[188] "radiotherapy_all_metastatic"
[189] "chemotherapy_number_lines_all_metastatic"
[190] "HER2_number_lines_all_metastatic"
[191] "endocrinetherapy_number_lines_all_metastatic"
[192] "treatment_other_all_metastatic"
[193] "date_last_update_file_database"
[194] "comments"
[195] "locoregional_recurrence"
[196] "date_locoregional_recurrence"
[197] "recurrence_contralateral_breast"
[198] "date_recurrence_contralateral_breast"
[199] "distant_recurrence"
[200] "date_distant_recurrence"
[201] "death"
[202] "date_of_death"
[203] "cause_of_death"
[204] "date_last_FU"
[205] "date_last_FU_Leuven" 2 DataCheckM0
2.1 Data-check
I loaded the data from the directory L:\\GBW-0080_BC_Lab\\Data\\FAT-ILC\\Giacomo
Table 3.1 reports the number of unknown values for each variable.
Table 3.2 shows the summary of the available information. skim_type, skim_variable, n_missing, complete_rate indicate the type of the variable, the name of the variable, the number of missing values and the proportion of complete values for each variable. Date.min, Date.max, Date.median, Date.n_unique indicate the minimum, maximum, the median and the number of unique values for the date variables. factor.n_unique, factor.top_counts indicate the number of unique values and the values with top counts for the categorical variables. numeric.p0, numeric.p25, numeric.p50, numeric.p75, numeric.p100 describe the percentiles of the numerical variable in the database.
As we will see later, several variables in the database result with a complete rate very low. The following variables have a complete rate of 0.
[1] "ER_H_score_biopsy, PR_H_score_biopsy, HER2_FISH_biopsy, HER2_ratio_biopsy, Ki67_biopsy, ER_H_score_biopsy_2nd_lesion, PR_H_score_biopsy_2nd_lesion, HER2_FISH_biopsy_2nd_lesion, HER2_ratio_biopsy_2nd_lesion, Ki67_biopsy_2nd_lesion, neo_adjuvant_chemotherapy_scheme, neo_adjuvant_chemotherapy_BSA_used, neo_adjuvant_chemotherapy_BSA_capping, neo_adjuvant_chemotherapy_completion, neo_adjuvant_HER2_therapy_scheme, neo_adjuvant_endocrinetherapy_scheme, neo_adjuvant_endocrinetherapy_duration, neo_adjuvant_other, residual_tumorbed, lobular_subtype, ER_H_score_resection_specimen_2nd_lesion, PR_H_score_resection_specimen_2nd_lesion, HER2_FISH_resection_specimen_2nd_lesion, HER2_ratio_resection_specimen_2nd_lesion, Ki67_resection_specimen_2nd_lesion, Antibody_E.cadherin, B.catenin, p120_catenin, GEP_outcome, adjuvant_HER2_scheme, chemotherapy_1st_line_metastatic, HER2_1st_line_metastatic, endocrinetherapy_1st_line_metastatic, treatment_1st_line_other_metastatic, treatment_reduction_1st_line_metastatic, clinical_response_1st_line_metastatic, chemotherapy_2nd_line_metastatic, HER2_2nd_line_metastatic, endocrinetherapy_2nd_line_metastatic, treatment_2nd_line_other_metastatic, treatment_reduction_2nd_line_metastatic, clinical_response_2nd_line_metastatic, second_progression_distant_disease_metastatic, radiotherapy_all_metastatic, chemotherapy_number_lines_all_metastatic, HER2_number_lines_all_metastatic, endocrinetherapy_number_lines_all_metastatic, treatment_other_all_metastatic, comments"The following variables have a complete rate above 0% but below 5%.
[1] "comorbidities, age_menarche, oral_anticonceptive_duration, fertility_treatment, age_last_pregnancy, breast_feeding, breast_feeding_duration, age_menopause, Number_of_adenopathies_expected_on_other_imaging, tumor_grade_biopsy, ER_Allred_biopsy, PR_Allred_biopsy, HER2_IHC_score_biopsy, ER_Allred_biopsy_2nd_lesion, PR_Allred_biopsy_2nd_lesion, HER2_IHC_score_biopsy_2nd_lesion, number_of_suspected_foci, Number_of_adenopathies_expected_on_imaging, ER_Allred_resection_specimen_2nd_lesion, PR_Allred_resection_specimen_2nd_lesion, HER2_IHC_score_resection_specimen_2nd_lesion, E.cadherin, lymphatic_invasion_resection_specimen, GEP_type, adjuvant_chemotherapy_scheme, adjuvant_endocrinetherapy_scheme1, adjuvant_endocrinetherapy_scheme1_duration, adjuvant_endocrinetherapy_scheme2, adjuvant_endocrinetherapy_scheme2_duration, Total_duration_endocrine_treatment, adjuvant_other"Instead, in ?tbl-skimsf are reported the variables that have a complete rate of at least 75%.
2.1.1 Missing values
Figure 3.1 displays in decreasing order the absolute frequency of the occurrence of missing values for each patient that has at least one missing value. For sake of simplicity, they are displayed separately depending on the number of the missing values. The same was performed for the variables, as displayed in Figure 3.2.
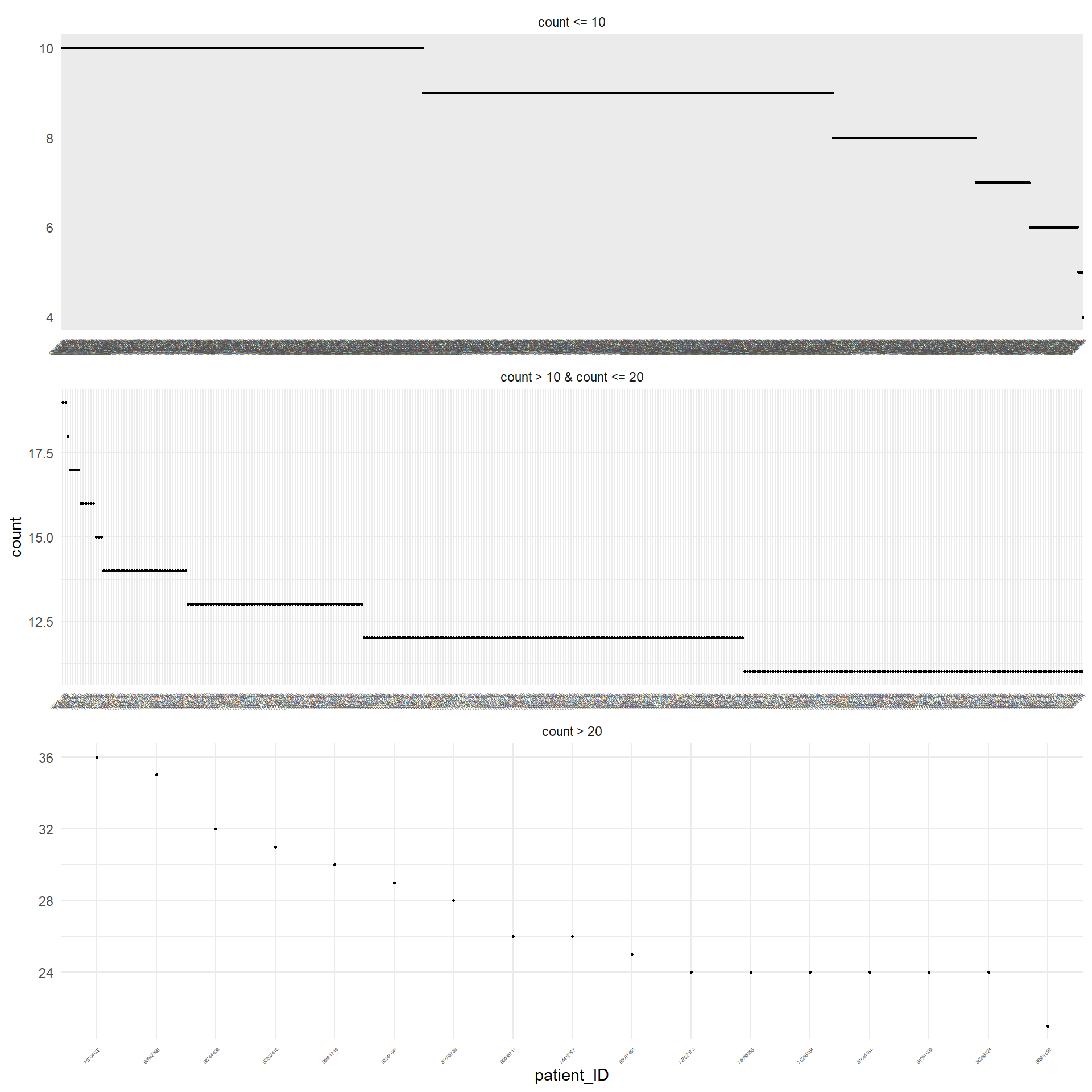
Table 3.3 reports the number of missing values for each patients.
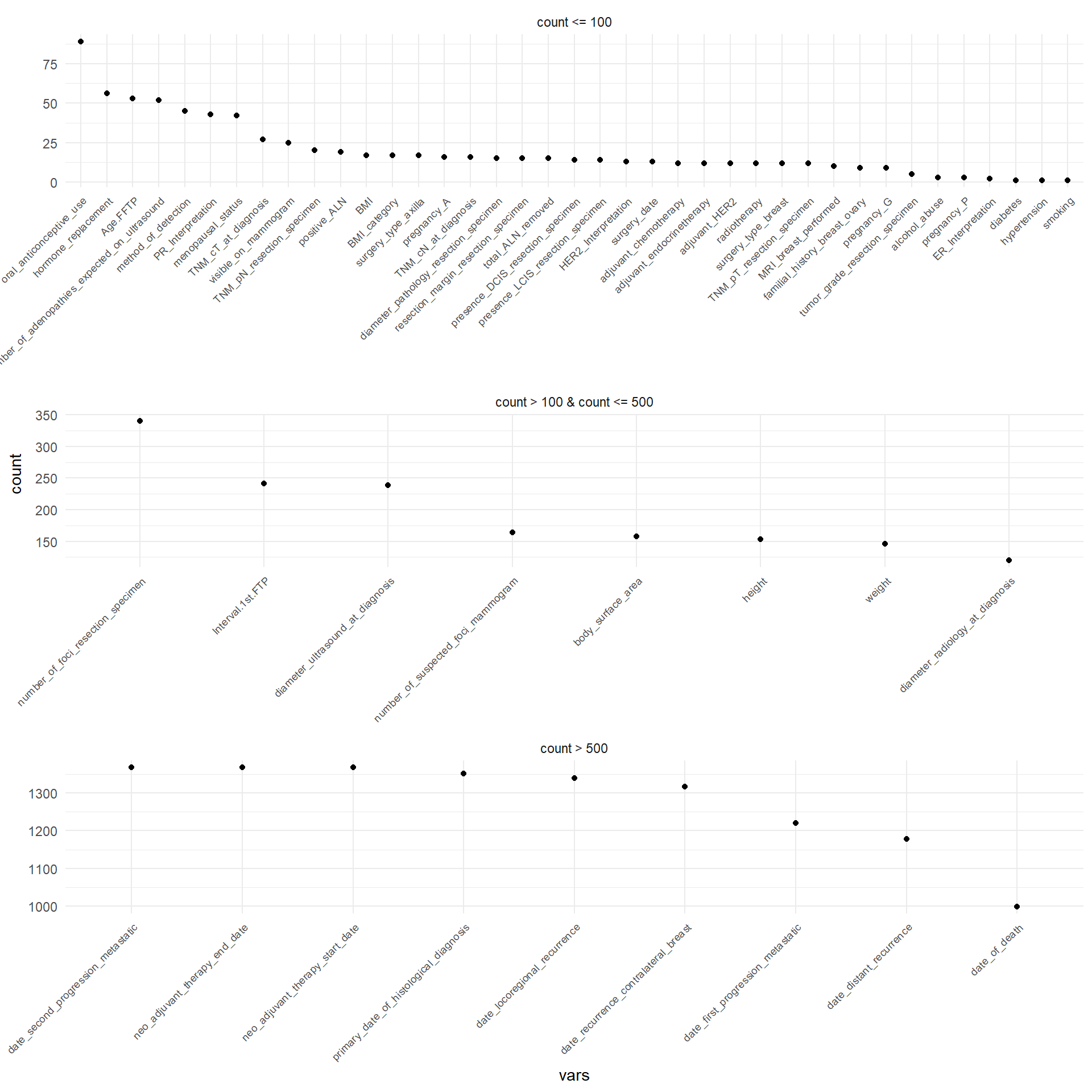
Table 3.4 reports the number of missing values for each variable.
2.1.2 Event history check
We need to set an order of event. The order is the following: Date of birth —> Date of diagnosis —> Date of start of NAT —> Date of end of NAT —> Surgery Date —> Date of recurrences —> Date first progression metastatic —–> Date second progression metastatic ——–> Date of death / Date of last FU / Date of last FU in own center.
Generally, the Date of last follow-up is equivalent to Date of death if death occurred. In some instances, Date of last follow-up is greater than Date of death. Why? Is Date of last follow-up referred to the day is known that the patients died at their respective Date of death?
Usually in the database, a Date of last follow-up in Leuven preceed Date of last follow-up at own center. Sometimes this is not the case, and Date of last follow-up in Leuven is greater than Date of last follow-up at own center. Is there a particular reason?
Then, some patients have a Date of last follow-up in Leuven that is before other events. This can be explained by the fact that the patients then are followed in their own centers.
There is one patient that has a Date of recurrence before the Date of Surgery.
Some patients do not have a date of surgery.
Some patients have a date of diagnosis equal to the date of surgery. How is it possible?
We have patients with multiple recurrence at different times. We will need to choose what is the most relevant type of recurrence. Is the distant the most relevant?
Patient 84323500 has a date of distant recurrence equal to a date of first progression.
# A tibble: 1 × 5
patient_ID date_distant_recurrence date_first_progression_metastatic
<chr> <date> <date>
1 84323500 2006-11-09 2006-11-09
# ℹ 2 more variables: date_recurrence_contralateral_breast <date>,
# date_locoregional_recurrence <date>2.1.3 Subset of variables : baseline characteristics
We now limit the analysis to the variables of interest. For the moment I will extract the following variables: method_of_detection, age_at_diagnosis, age_category, BMI, BMI_category, menopausal_status, smoking, alcohol_abuse, hypertension, hyperlipidemia, diabetes, oral_anticonceptive_use, pregnancy_P, hormone_replacement, germline_mutation_testing_performed, germline_mutation_testing_result,germline_mutation_testing_year_most_recent_test, familial_history_breast_ovary, visible_on_mammogram, TNM_cT_at_diagnosis, TNM_cN_at_diagnosis, TNM_cM_at_diagnosis, neo_adjuvant_therapy, surgery_type_breast, surgery_type_axilla, TNM_pT_resection_specimen, TNM_pN_resection_specimen, diameter_pathology_resection_specimen, tumor_grade_resection_specimen, resection_margin_resection_specimen, ER_Interpretation, PR_Interpretation, HER2_Interpretation, presence_DCIS_resection_specimen, presence_LCIS_resection_specimen, radiotherapy, adjuvant_chemotherapy, adjuvant_HER2, adjuvant_endocrinetherapy, multifocality,.
Table 3.5 reports the first description of the variables included in the analysis.
[1] "patient_ID+ method_of_detection+ age_at_diagnosis+ age_category+ BMI+ BMI_category+ menopausal_status+ smoking+ alcohol_abuse+ hypertension+ hyperlipidemia+ diabetes+ oral_anticonceptive_use+ pregnancy_A+ pregnancy_P+ hormone_replacement+ familial_history_breast_ovary+ visible_on_mammogram+ TNM_cT_at_diagnosis+ TNM_cN_at_diagnosis+ TNM_cM_at_diagnosis+ neo_adjuvant_therapy+ surgery_type_breast+ surgery_type_axilla+ TNM_pT_resection_specimen+ TNM_pN_resection_specimen+ diameter_pathology_resection_specimen+ tumor_grade_resection_specimen+ resection_margin_resection_specimen+ ER_Interpretation+ PR_Interpretation+ HER2_Interpretation+ presence_DCIS_resection_specimen+ presence_LCIS_resection_specimen+ radiotherapy+ adjuvant_chemotherapy+ adjuvant_HER2+ adjuvant_endocrinetherapy+ germline_mutation_testing_performed+ germline_mutation_testing_result+ germline_mutation_testing_year_most_recent_test+ multifocality+ meta_brain_nonleptomeningeal_atfirstprogression+ meta_leptomeningeal_atfirstprogression+ meta_bones_atfirstprogression+ meta_skin_atfirstprogression+ meta_lungs_atfirstprogression+ meta_liver_atfirstprogression+ meta_abdomen_extrahepatic_atfirstprogression+ meta_reproductive_organs_atfirstprogression+ meta_lymph_nodes_atfirstprogression+ meta_other_atfirstprogression"| Overall (N=1367) |
|
|---|---|
| method_of_detection | |
| radiologically detected | 548 (40.1%) |
| symptoms | 773 (56.5%) |
| Missing | 46 (3.4%) |
| age_at_diagnosis | |
| Mean (SD) | 61.5 (11.8) |
| Median [Min, Max] | 61.0 [32.0, 95.0] |
| Missing | 1 (0.1%) |
| age_category | |
| < 40 | 23 (1.7%) |
| ≥ 80 | 106 (7.8%) |
| 40 - 49 | 209 (15.3%) |
| 50 - 59 | 387 (28.3%) |
| 60 - 69 | 397 (29.0%) |
| 70 - 79 | 244 (17.8%) |
| Missing | 1 (0.1%) |
| BMI | |
| Mean (SD) | 25.6 (4.85) |
| Median [Min, Max] | 24.8 [14.9, 47.7] |
| Missing | 18 (1.3%) |
| BMI_category | |
| < 18,5 | 29 (2.1%) |
| ≥18,5 and <25 | 674 (49.3%) |
| ≥25 and <30 | 424 (31.0%) |
| ≥30 | 222 (16.2%) |
| Missing | 18 (1.3%) |
| menopausal_status | |
| Postmenopausal | 981 (71.8%) |
| pre- and perimenopausal | 343 (25.1%) |
| Missing | 43 (3.1%) |
| smoking | |
| active | 188 (13.8%) |
| former | 266 (19.5%) |
| no | 911 (66.6%) |
| Missing | 2 (0.1%) |
| alcohol_abuse | |
| no | 1158 (84.7%) |
| yes | 205 (15.0%) |
| Missing | 4 (0.3%) |
| hypertension | |
| no | 843 (61.7%) |
| yes | 522 (38.2%) |
| Missing | 2 (0.1%) |
| hyperlipidemia | |
| no | 1058 (77.4%) |
| yes | 308 (22.5%) |
| Missing | 1 (0.1%) |
| diabetes | |
| MODY | 1 (0.1%) |
| no | 1281 (93.7%) |
| type 1 | 3 (0.2%) |
| type 2 | 80 (5.9%) |
| Missing | 2 (0.1%) |
| oral_anticonceptive_use | |
| active | 180 (13.2%) |
| former | 670 (49.0%) |
| no | 427 (31.2%) |
| Missing | 90 (6.6%) |
| pregnancy_A | |
| 0 | 1052 (77.0%) |
| 1 | 204 (14.9%) |
| 10 | 1 (0.1%) |
| 2 | 64 (4.7%) |
| 3 | 18 (1.3%) |
| 4 | 8 (0.6%) |
| 5 | 2 (0.1%) |
| 6 | 1 (0.1%) |
| Missing | 17 (1.2%) |
| pregnancy_P | |
| 0 | 188 (13.8%) |
| 1 | 293 (21.4%) |
| 10 | 2 (0.1%) |
| 12 | 1 (0.1%) |
| 2 | 529 (38.7%) |
| 3 | 231 (16.9%) |
| 4 | 83 (6.1%) |
| 5 | 25 (1.8%) |
| 6 | 8 (0.6%) |
| 9 | 3 (0.2%) |
| Missing | 4 (0.3%) |
| hormone_replacement | |
| active | 204 (14.9%) |
| former | 167 (12.2%) |
| no | 939 (68.7%) |
| Missing | 57 (4.2%) |
| familial_history_breast_ovary | |
| no | 825 (60.4%) |
| yes | 532 (38.9%) |
| Missing | 10 (0.7%) |
| visible_on_mammogram | |
| no | 123 (9.0%) |
| yes | 1219 (89.2%) |
| Missing | 25 (1.8%) |
| TNM_cT_at_diagnosis | |
| T1a | 13 (1.0%) |
| T1b | 140 (10.2%) |
| T1c | 357 (26.1%) |
| T1mi | 1 (0.1%) |
| T2 | 594 (43.5%) |
| T3 | 193 (14.1%) |
| T4a | 3 (0.2%) |
| T4b | 37 (2.7%) |
| T4c | 2 (0.1%) |
| T4d | 10 (0.7%) |
| Tis | 10 (0.7%) |
| Missing | 7 (0.5%) |
| TNM_cN_at_diagnosis | |
| N0 | 1124 (82.2%) |
| N1 | 195 (14.3%) |
| N2 | 11 (0.8%) |
| N3a | 18 (1.3%) |
| N3b | 3 (0.2%) |
| N3c | 6 (0.4%) |
| Missing | 10 (0.7%) |
| TNM_cM_at_diagnosis | |
| M0 | 1366 (99.9%) |
| Missing | 1 (0.1%) |
| neo_adjuvant_therapy | |
| no | 1256 (91.9%) |
| yes: anti-HER2 ADC + trastuzumab + pertuzumab | 1 (0.1%) |
| yes: CT | 39 (2.9%) |
| yes: CT + ET | 2 (0.1%) |
| yes: CT + ET + trastuzumab + pertuzumab | 2 (0.1%) |
| yes: CT + ICI | 1 (0.1%) |
| yes: CT + trastuzumab | 11 (0.8%) |
| yes: CT + trastuzumab + pertuzumab | 3 (0.2%) |
| yes: ET | 42 (3.1%) |
| yes: ET + CDK4/6i | 3 (0.2%) |
| yes: ET + ROSi | 6 (0.4%) |
| Missing | 1 (0.1%) |
| surgery_type_breast | |
| Mastectomy | 810 (59.3%) |
| Tumorectomy | 517 (37.8%) |
| Tumorectomy + Mastectomy | 27 (2.0%) |
| Missing | 13 (1.0%) |
| surgery_type_axilla | |
| ALN | 522 (38.2%) |
| SLN | 679 (49.7%) |
| SLN + ALN | 148 (10.8%) |
| Missing | 18 (1.3%) |
| TNM_pT_resection_specimen | |
| T0 | 10 (0.7%) |
| T1a | 23 (1.7%) |
| T1b | 98 (7.2%) |
| T1c | 327 (23.9%) |
| T1mi | 1 (0.1%) |
| T2 | 570 (41.7%) |
| T3 | 318 (23.3%) |
| T4b | 7 (0.5%) |
| Missing | 13 (1.0%) |
| TNM_pN_resection_specimen | |
| N0(i-) | 727 (53.2%) |
| N0(i+) | 80 (5.9%) |
| N1a | 271 (19.8%) |
| N1mi | 77 (5.6%) |
| N2a | 88 (6.4%) |
| N3a | 102 (7.5%) |
| N3b | 1 (0.1%) |
| Missing | 21 (1.5%) |
| diameter_pathology_resection_specimen | |
| Mean (SD) | 38.5 (29.7) |
| Median [Min, Max] | 30.0 [0, 220] |
| Missing | 16 (1.2%) |
| tumor_grade_resection_specimen | |
| 1 | 16 (1.2%) |
| 2 | 1211 (88.6%) |
| 3 | 134 (9.8%) |
| Missing | 6 (0.4%) |
| resection_margin_resection_specimen | |
| dubious (< 1 mm) | 145 (10.6%) |
| negative | 1132 (82.8%) |
| positive | 74 (5.4%) |
| Missing | 16 (1.2%) |
| ER_Interpretation | |
| negative | 25 (1.8%) |
| positive | 1339 (98.0%) |
| Missing | 3 (0.2%) |
| PR_Interpretation | |
| negative | 169 (12.4%) |
| positive | 1154 (84.4%) |
| Missing | 44 (3.2%) |
| HER2_Interpretation | |
| negative | 1294 (94.7%) |
| positive | 59 (4.3%) |
| Missing | 14 (1.0%) |
| presence_DCIS_resection_specimen | |
| no | 1198 (87.6%) |
| yes | 154 (11.3%) |
| Missing | 15 (1.1%) |
| presence_LCIS_resection_specimen | |
| no | 227 (16.6%) |
| yes, classical LCIS | 770 (56.3%) |
| yes, non classical LCIS | 355 (26.0%) |
| Missing | 15 (1.1%) |
| radiotherapy | |
| no | 238 (17.4%) |
| yes | 1116 (81.6%) |
| Missing | 13 (1.0%) |
| adjuvant_chemotherapy | |
| no | 1005 (73.5%) |
| yes | 349 (25.5%) |
| Missing | 13 (1.0%) |
| adjuvant_HER2 | |
| no | 1315 (96.2%) |
| yes | 39 (2.9%) |
| Missing | 13 (1.0%) |
| adjuvant_endocrinetherapy | |
| no | 46 (3.4%) |
| yes | 1308 (95.7%) |
| Missing | 13 (1.0%) |
| germline_mutation_testing_performed | |
| no | 1070 (78.3%) |
| yes | 296 (21.7%) |
| Missing | 1 (0.1%) |
| germline_mutation_testing_result | |
| ATM | 2 (0.1%) |
| BRCA1 | 1 (0.1%) |
| BRCA2 | 11 (0.8%) |
| CDH1 | 2 (0.1%) |
| CHEK2 | 2 (0.1%) |
| MSH6 | 1 (0.1%) |
| negative | 277 (20.3%) |
| ongoing | 2 (0.1%) |
| PALB2 | 2 (0.1%) |
| Missing | 1067 (78.1%) |
| germline_mutation_testing_year_most_recent_test | |
| Mean (SD) | 2020 (4.99) |
| Median [Min, Max] | 2020 [2000, 2030] |
| Missing | 1069 (78.2%) |
| multifocality | |
| multifocal | 285 (20.8%) |
| unifocal | 1057 (77.3%) |
| Missing | 25 (1.8%) |
| meta_brain_nonleptomeningeal_atfirstprogression | |
| NA | 1214 (88.8%) |
| no | 136 (9.9%) |
| unknown | 8 (0.6%) |
| yes | 8 (0.6%) |
| Missing | 1 (0.1%) |
| meta_leptomeningeal_atfirstprogression | |
| NA | 1214 (88.8%) |
| no | 137 (10.0%) |
| unknown | 8 (0.6%) |
| yes | 7 (0.5%) |
| Missing | 1 (0.1%) |
| meta_bones_atfirstprogression | |
| NA | 1214 (88.8%) |
| no | 69 (5.0%) |
| unknown | 8 (0.6%) |
| yes | 75 (5.5%) |
| Missing | 1 (0.1%) |
| meta_skin_atfirstprogression | |
| NA | 1214 (88.8%) |
| no | 137 (10.0%) |
| unknown | 8 (0.6%) |
| yes | 7 (0.5%) |
| Missing | 1 (0.1%) |
| meta_lungs_atfirstprogression | |
| NA | 1214 (88.8%) |
| no | 139 (10.2%) |
| unknown | 8 (0.6%) |
| yes | 5 (0.4%) |
| Missing | 1 (0.1%) |
| meta_liver_atfirstprogression | |
| NA | 1214 (88.8%) |
| no | 104 (7.6%) |
| unknown | 8 (0.6%) |
| yes | 40 (2.9%) |
| Missing | 1 (0.1%) |
| meta_abdomen_extrahepatic_atfirstprogression | |
| NA | 1214 (88.8%) |
| no | 98 (7.2%) |
| unknown | 8 (0.6%) |
| yes | 46 (3.4%) |
| Missing | 1 (0.1%) |
| meta_reproductive_organs_atfirstprogression | |
| NA | 1214 (88.8%) |
| no | 139 (10.2%) |
| unknown | 8 (0.6%) |
| yes | 5 (0.4%) |
| Missing | 1 (0.1%) |
| meta_lymph_nodes_atfirstprogression | |
| NA | 1214 (88.8%) |
| no | 115 (8.4%) |
| unknown | 8 (0.6%) |
| yes | 29 (2.1%) |
| Missing | 1 (0.1%) |
| meta_other_atfirstprogression | |
| NA | 1214 (88.8%) |
| no | 107 (7.8%) |
| unknown | 7 (0.5%) |
| yes: adrenal | 2 (0.1%) |
| yes: adrenal and pleura | 1 (0.1%) |
| yes: adrenal and retroperitoneal | 1 (0.1%) |
| yes: biochemical | 5 (0.4%) |
| yes: bladder | 2 (0.1%) |
| yes: kidney | 1 (0.1%) |
| yes: muscle | 1 (0.1%) |
| yes: orbit | 1 (0.1%) |
| yes: pericard and pleura | 1 (0.1%) |
| yes: pleura | 15 (1.1%) |
| yes: pleura and muscle | 1 (0.1%) |
| yes: pleura and pericard | 1 (0.1%) |
| yes: retroperitoneal | 5 (0.4%) |
| yes: retroperitoneal and fat | 1 (0.1%) |
| Missing | 1 (0.1%) |
2.1.4 Number of events
I am excluding for the moment those patients that have a date of surgery equal to the date of diagnosis and all the other patients whose dates were unsure.
Figure 3.3 describes the event history of the patients. You just need to pick a starting state from the ‘from’ axis and select then transitioning state from the ‘to’ axis. In the corresponding cell, you find the absolute frequency of each transition.
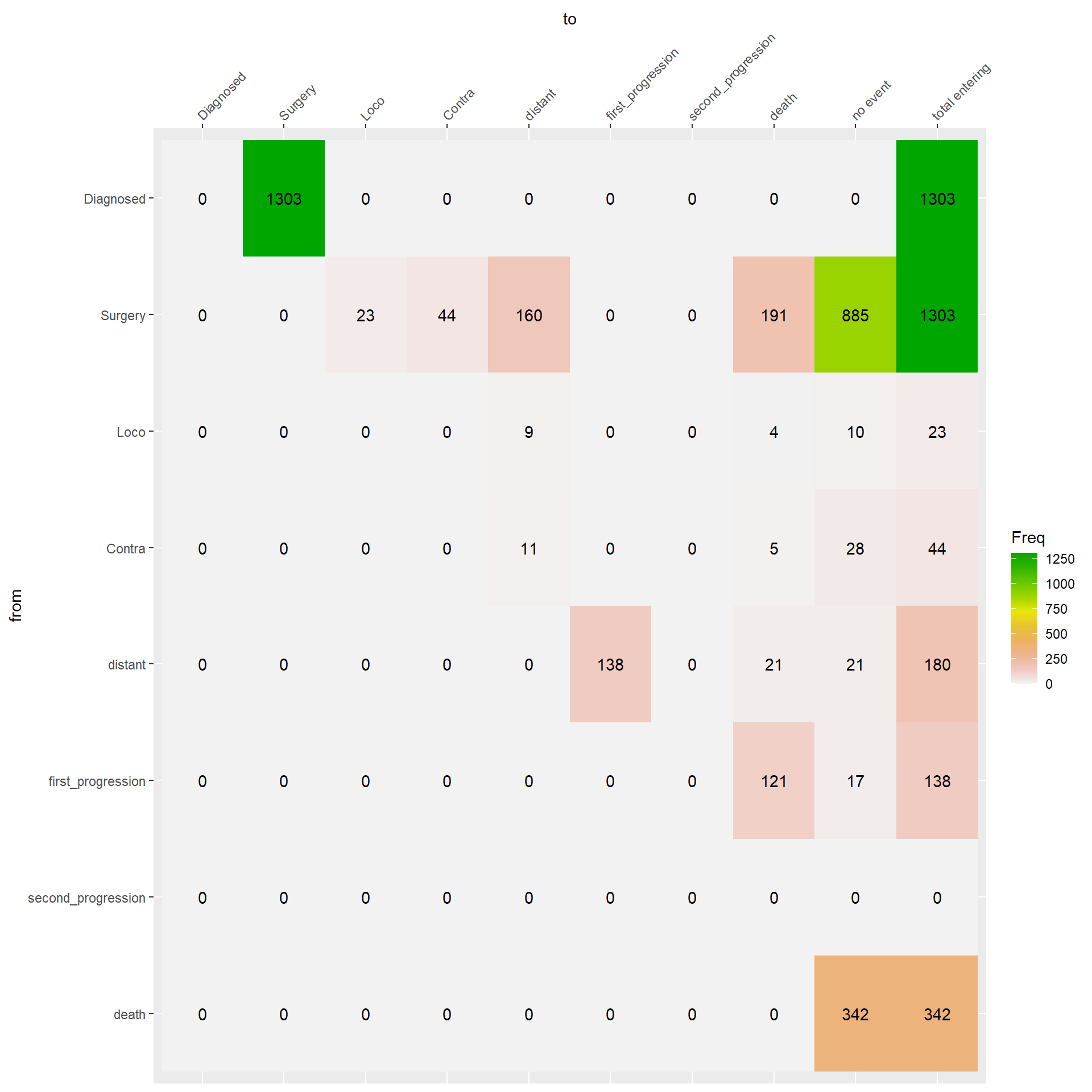
Check the patients with a date of lost to follow-up before date of death
Look at the distribution of the variables through the years
2.1.5 Check patients lost to follow-up before death.
This dotplot shows the distribution of the values of the difference in days between the day of death and the day of last follow-up, for the patients who had a date of last follow-up before the date of death.
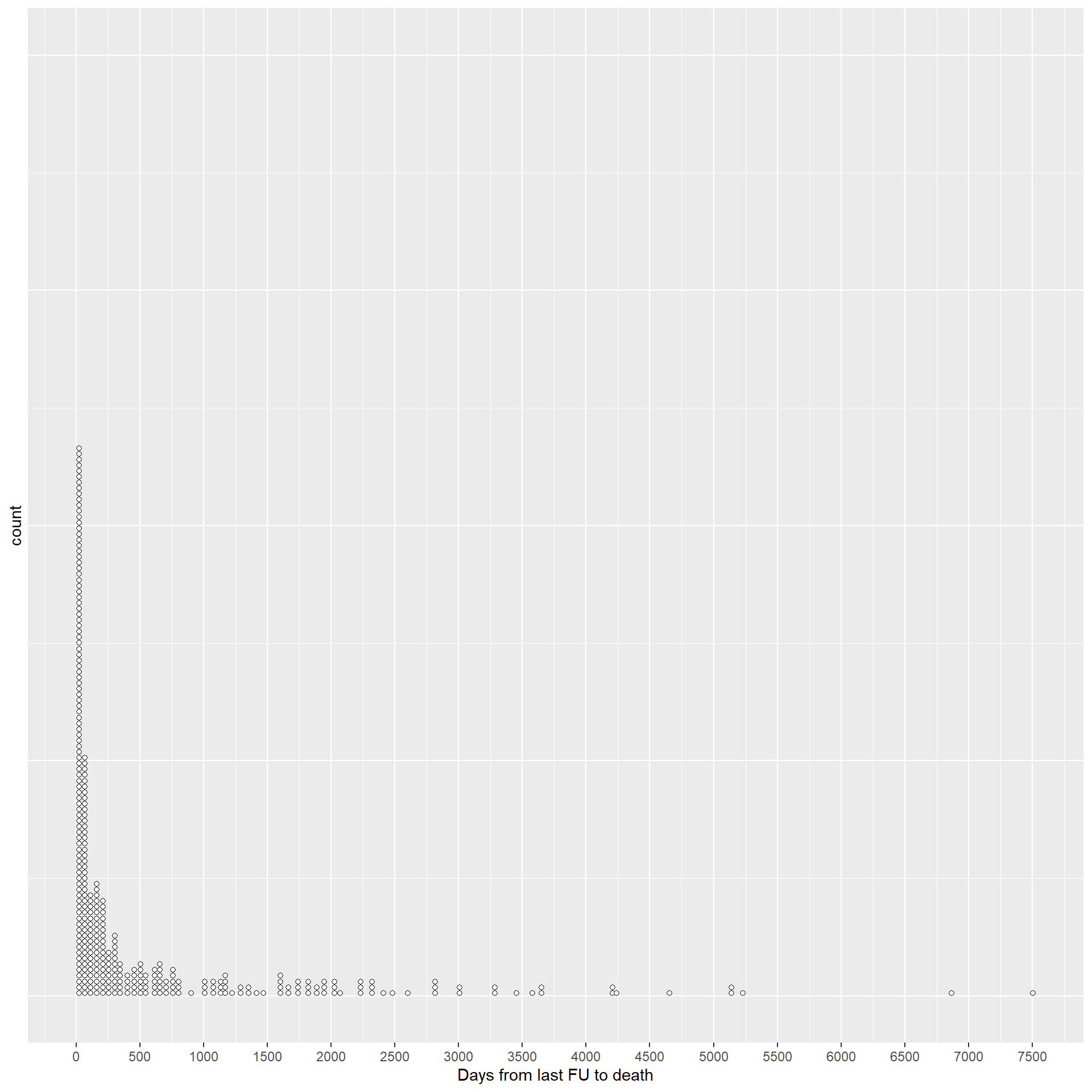
The table reports the patients who died after more than 1 year from their last day of follow-up.